12 Bed File Tips For Easy Bam Conversion

The conversion of BED (Browser Extensible Data) files to BAM (Binary Alignment/Map) format is a common task in bioinformatics and genomics research. BED files are used to represent genomic regions of interest, while BAM files are used for storing large amounts of sequencing data in a compressed, binary format. The conversion between these two formats can be crucial for various analyses, such as genome assembly, variant detection, and gene expression studies. Here, we provide 12 tips for easy BAM conversion from BED files, focusing on practical steps, tools, and considerations to ensure efficient and accurate conversion.
Understanding BED and BAM Formats
Before diving into the conversion process, it’s essential to understand the structure and content of both BED and BAM files. BED files are plain text files that contain information about genomic regions, including the chromosome, start position, end position, and sometimes additional features like gene names or scores. BAM files, on the other hand, are binary files that store aligned sequencing reads against a reference genome, including information about the read, its mapping quality, and any mismatches or gaps.
The conversion from BED to BAM typically involves aligning sequencing reads to the reference genome and then filtering or selecting reads based on the regions of interest defined in the BED file. Samtools and BEDTools are popular software suites used for manipulating and analyzing these file formats.
Preparation of BED Files
Ensuring that BED files are correctly formatted and contain accurate genomic coordinates is crucial for successful conversion and subsequent analysis. The bed format can vary (e.g., bed3, bed6, bed12), with each specifying the minimum number of fields required per line. A standard bed3 file, for example, would contain the chromosome name, start position (0-based), and end position (1-based) for each feature. Chromosomal coordinates must be correctly specified, considering the reference genome version used.
A sample bed3 file might look like this:
| Chromosome | Start | End |
|---|---|---|
| chr1 | 10000 | 20000 |
| chr2 | 50000 | 60000 |
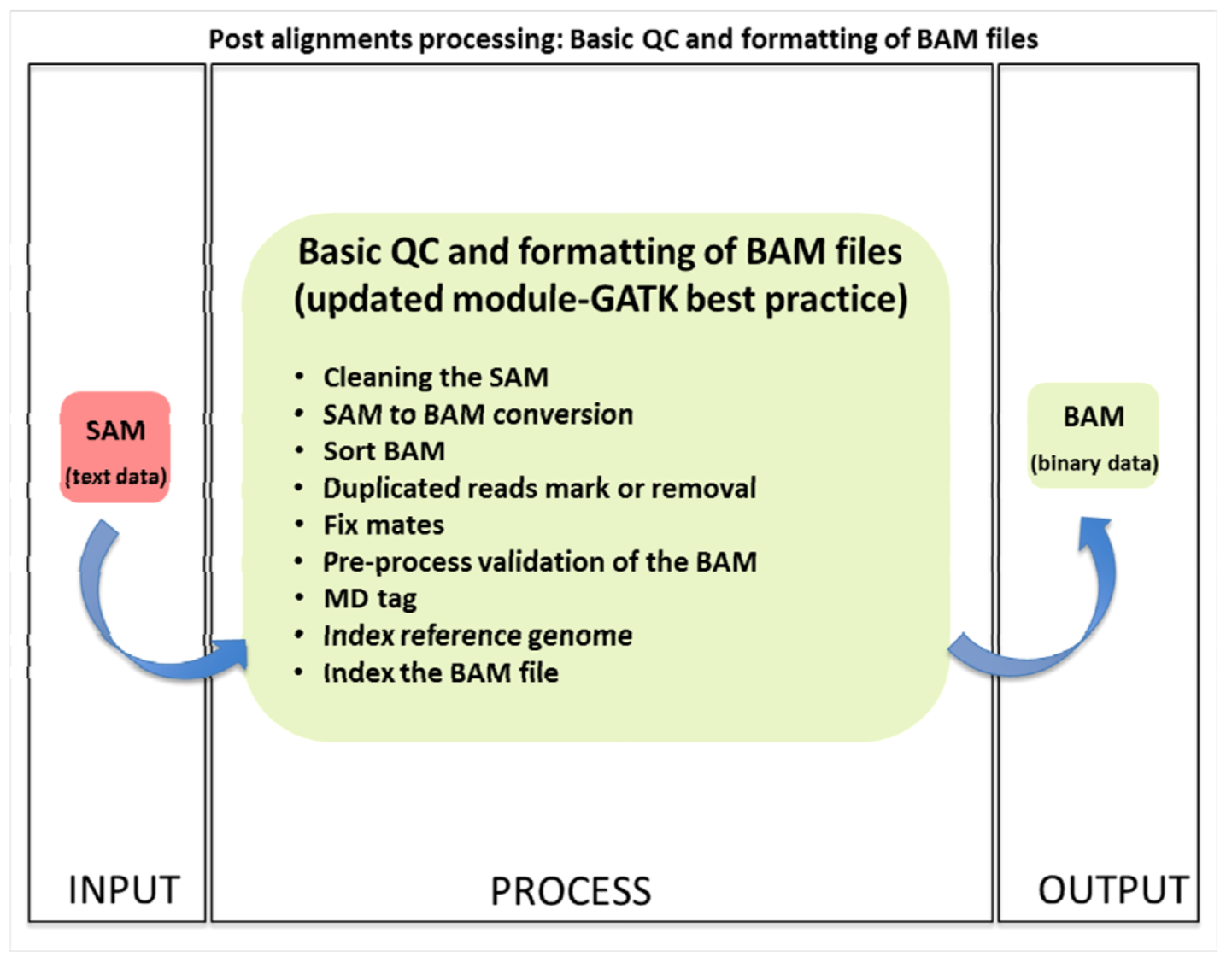
Conversion Tools and Commands

Several tools can be used for the conversion and analysis process, including BEDTools, Samtools, and BWA (Burrows-Wheeler Aligner). For example, to intersect a BAM file with a BED file and output the overlapping reads, you might use the bedtools intersect command. If aligning reads to a reference genome is necessary, bwa mem followed by samtools view and samtools sort can be used to generate a sorted BAM file.
The choice of tool and specific command-line options depend on the analysis goals, the nature of the sequencing data, and the computational resources available. Optimizing these commands for performance, especially when dealing with large datasets, is crucial.
Performance Considerations
Converting and analyzing large genomic datasets can be computationally intensive. Parallel processing and the use of high-performance computing (HPC) environments can significantly speed up the analysis. Additionally, optimizing disk usage by ensuring sufficient storage space and using efficient file formats can prevent bottlenecks during the conversion and analysis process.
For large-scale analyses, it might be beneficial to use cloud computing platforms or containerization tools like Docker to ensure reproducibility and scalability of the workflow.
What is the purpose of converting BED files to BAM format?
+The conversion allows for the integration of genomic region information with aligned sequencing data, facilitating various downstream analyses such as the identification of variants within specific regions of interest.
How do I ensure the accuracy of genomic coordinates in my BED file?
+Verify the reference genome version used and cross-check coordinates against reliable genomic databases or annotation files to ensure accuracy and consistency.
In conclusion, the conversion of BED files to BAM format for genomic analysis requires careful preparation of the input files, selection of appropriate tools and commands, and consideration of computational performance. By following these 12 tips and staying informed about the latest developments in bioinformatics tools and methodologies, researchers can efficiently and accurately convert and analyze their genomic data, ultimately contributing to a deeper understanding of the genome and its functions.
